Alzheimer's disease
Genomics approaches to study the molecular and cellular aspects of Alzheimer's disease.
Much of my PhD research in the Swarup Lab has focused on studying Alzheimer’s Disease (AD) and neurodegeneration using sequencing approaches like single-cell and spatial -omics. Here I briefly describe each of these projects in chronological order, encompassing my overall PhD journey.
1. Integrative genomics approach identifies conserved transcriptomic networks in Alzheimer’s disease
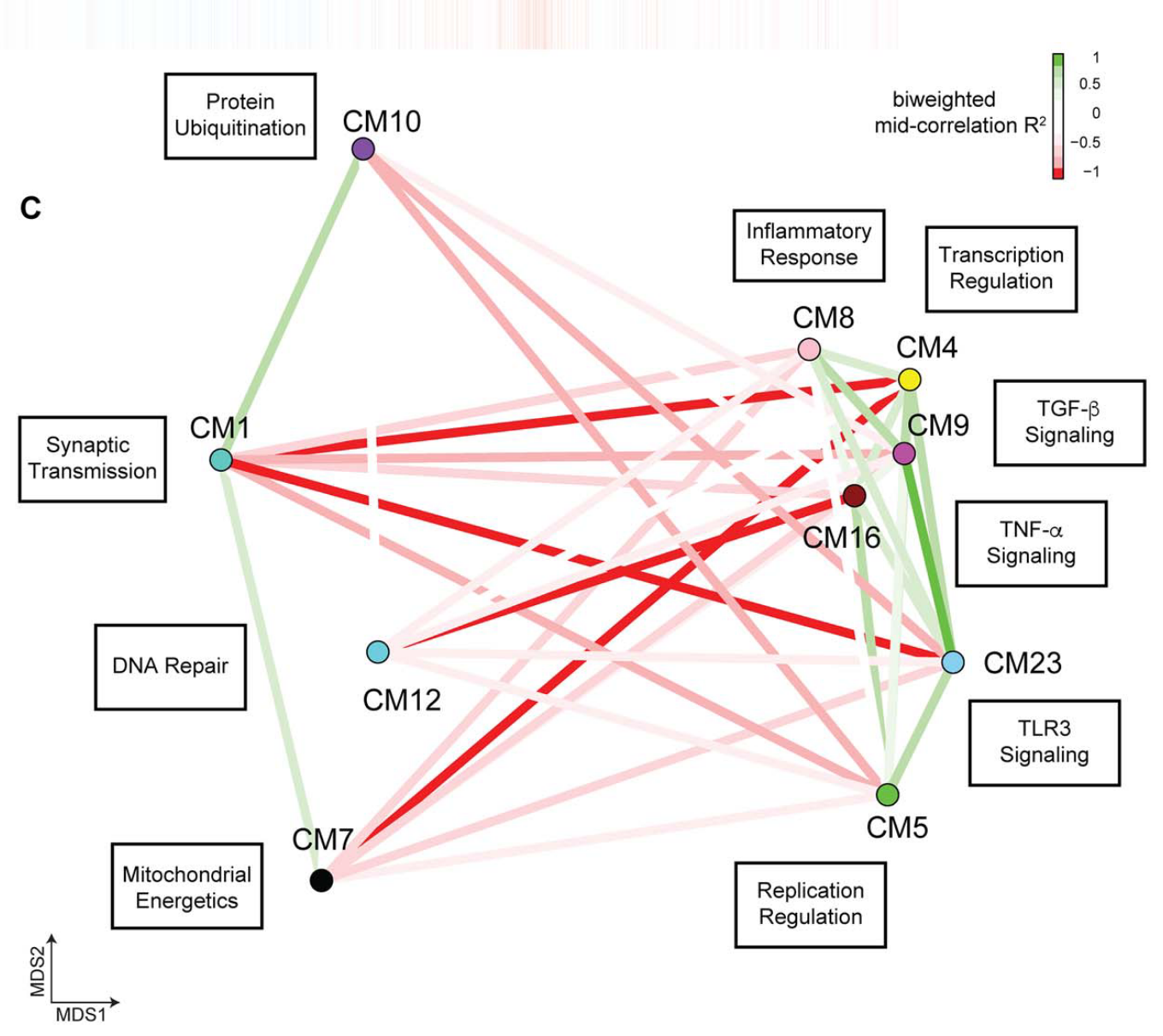
In this project, we collected over 1,200 bulk RNA-seq samples from AD brains using three publicly available datasets (ROSMAP, Mt. Sinai Medical, and Mayo Clinic). We performed consensus weighted gene co-expression network analysis (cWGCNA) across these different datasets and brain regions, identifying several gene modules that were altered in disease. This project started out as my rotation project in the Swarup Lab during first year of my PhD, and it eventually became my first first-author publication (Morabito et al., 2020).
2. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease
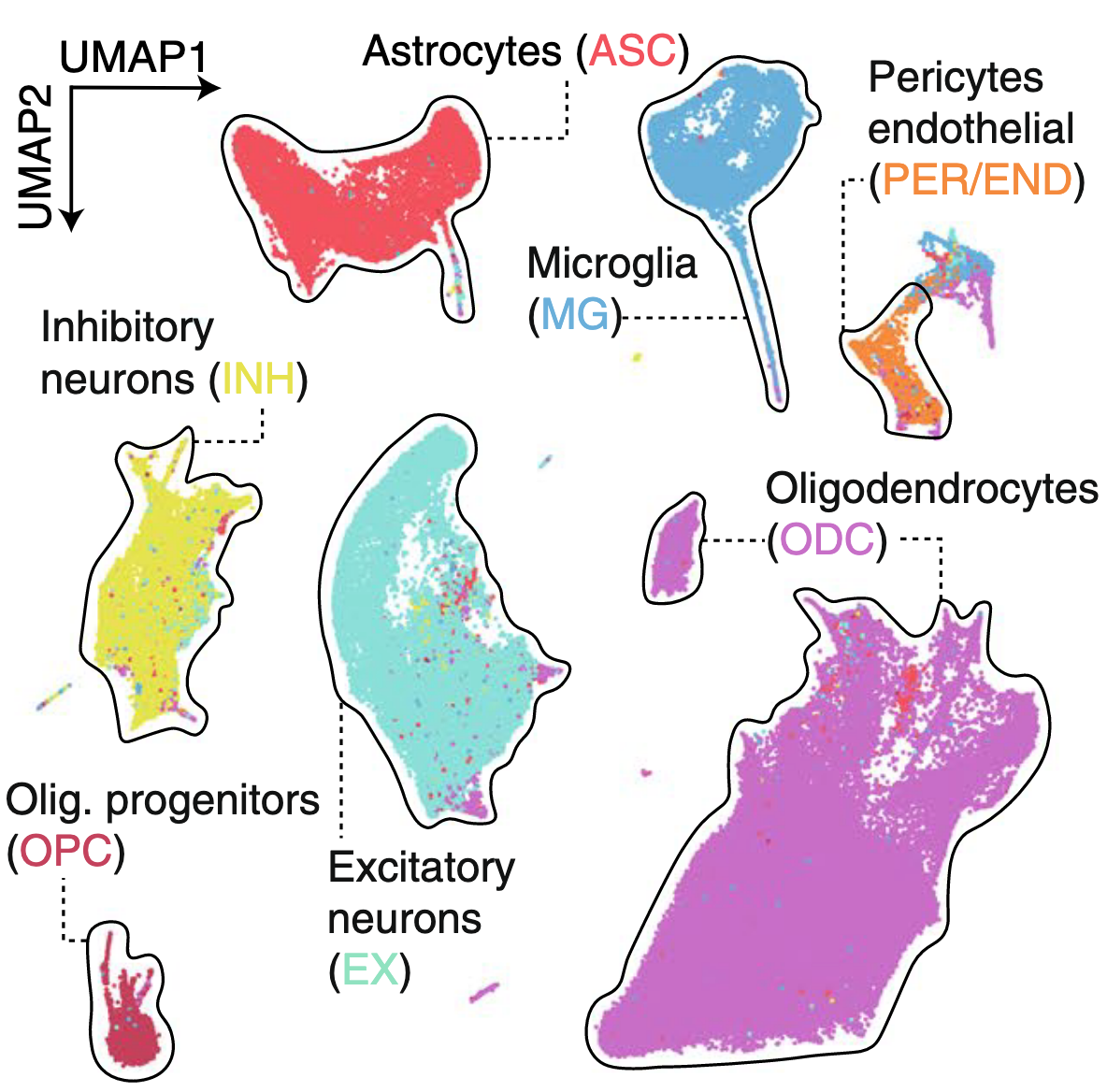
This work was the first of its kind to profile the epigenome and transcriptome in AD brains in single cells (Morabito* et al., 2021). In 2019, we began to generate snATAC-seq and snRNA-seq libraries in prefrontal cortex samples of AD donors. We characterized differences in the epigenome and transcriptome between AD and cognitively normal controls for each brain cell type. We used our multi-omic information to link cis-regulatory elements to candidate target genes, and investigated cell-type specific epigenomic landscapes at genetic risk loci. Much of my work on the computational side of this project took place during the onset of the COVID-19 pandemic.
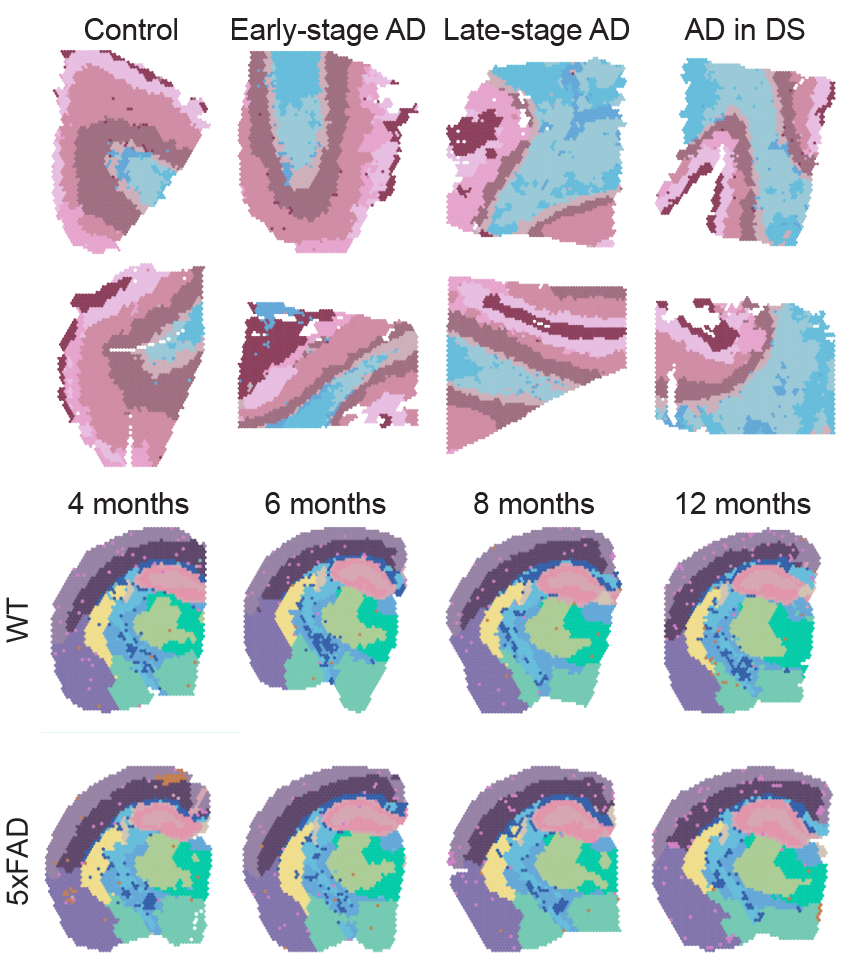
3. Spatial and single-nucleus transcriptomic analysis of genetic and sporadic forms of Alzheimer’s Disease
Here we applied spatial and single-nucleus transcriptomics to compare genetic and sporadic forms of AD (Miyoshi* et al., 2023). In particular, we focused comparing Down Syndrome individuals, who generally develop AD with aging (AD in DS), with the sporadic AD population. We also generated spatial data in the 5xFAD mouse model for further comparisons and to identify amyloid-associated gene signatures with aging. We uncovered genes and networks which are similar between AD in DS and sporadic AD, and some which are specific to certain disease stages. Multi-scale network analysis identified an inflammatory glial gene network that we linked to AD polygenic risk and to amyloid accumulation. This project represents the second half of my PhD research.