network analysis
Developing and applying tools for network analysis in high-dimensional -omics data.
Network analysis in genomic data is one of my primary research interests. Biological systems are highly complex and intrinsically interconnected, and there is much we can learn about diseases and other conditions by interrogating genomic networks. Many of my research projects have involved applying existing network analysis tools, but I have also worked on developing such tools like hdWGCNA.
Method development
One of my primary research interests is in method development for network analysis in genomics data. I am the lead developer of hdWGCNA, an R package for co-expression network analysis in single-cell and spatial transcriptomics data. This method constructs networks in specific contexts that reflect the underlying biology of individual cell types or disease conditions. hdWGCNA also includes downstream tools for data visualization and statistical testing. We developed this package to be user-friendly and we leveraged common data formats which enable a borad range of researchers to use this package. To date hdWGCNA has already gathered hundreds of users across many different fields of research. Check out our publication to learn more about this method (Morabito et al., 2023).

Applications
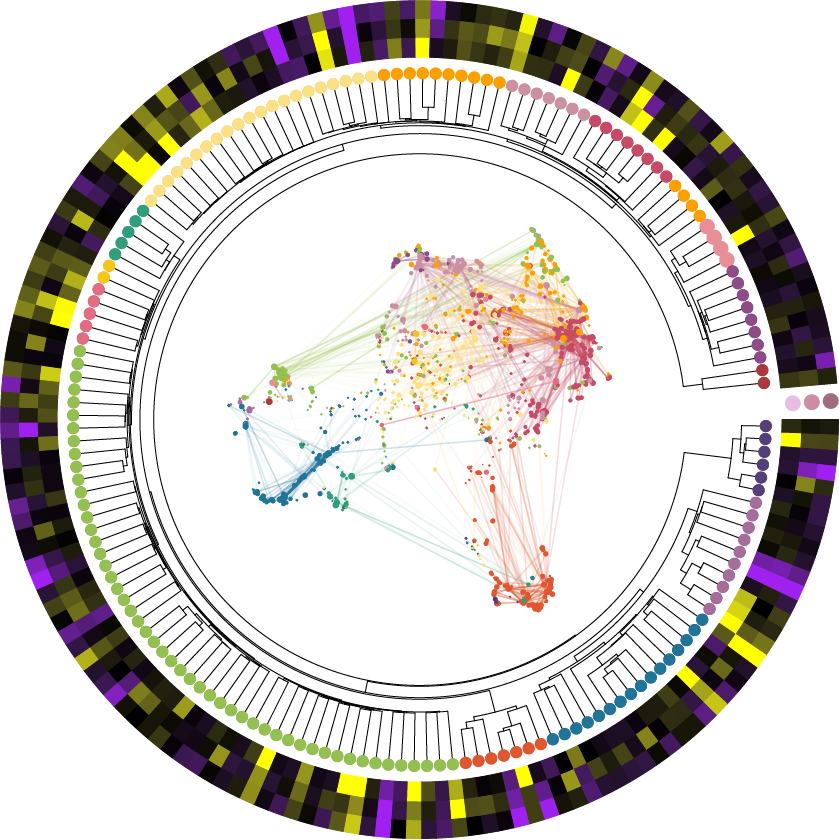
Nearly all of my research projects have involved applying network analysis approaches in some capacity. These approaches enable us to identify systems-level changes in disease or in specific cell states, beyond what differential expression testing is capable of revealing. For example, in our recent preprint (Miyoshi* et al., 2023) we performed multi-scale network analysis across different cortical layers in our spatial dataset to reveal an altered neuroinflammatory signature in disease. In some cases, I have also worked on other types of network analysis aside from co-expression networks, like transcription factor regulatory networks in (Morabito* et al., 2021).

