publications
2025
-
 Single-nucleus multi-omics identifies shared and distinct pathways in Pick’s and Alzheimer’s diseaseZechuan Shi, Sudeshna Das, Samuel Morabito, and 12 more authorsScience Advances, 2025
Single-nucleus multi-omics identifies shared and distinct pathways in Pick’s and Alzheimer’s diseaseZechuan Shi, Sudeshna Das, Samuel Morabito, and 12 more authorsScience Advances, 2025The study of transcriptomic and epigenomic variations in neurodegenerative diseases, particularly tauopathies like Pick’s disease (PiD) and Alzheimer’s disease (AD), offers insights into their underlying regulatory mechanisms. Here, we identified critical regulatory changes driving disease progression, revealing potential therapeutic targets. Our comparative analyses uncovered disease-enriched noncoding regions and genome-wide transcription factor (TF) binding differences, linking them to target genes. Notably, we identified a distal human-gained enhancer (HGE) associated with E3 ubiquitin ligase (UBE3A), highlighting disease-specific regulatory alterations. Additionally, fine mapping of AD risk genes uncovered loci enriched in microglial enhancers and accessible in other cell types. Shared and distinct TF binding patterns were observed in neurons and glial cells across PiD and AD. We validated our findings using CRISPR to excise a predicted enhancer region in UBE3A and developed an interactive database, scROAD, to visualize predicted single-cell TF occupancy and regulatory networks.
-
 Genetically determined Alzheimer’s disease research advances: The Down Syndrome & Autosomal Dominant Alzheimer’s Disease 2024 ConferenceNeus Falgàs, Lucia Maure‐Blesa, Beau Ances, and 56 more authorsAlzheimer’s & Dementia, 2025
Genetically determined Alzheimer’s disease research advances: The Down Syndrome & Autosomal Dominant Alzheimer’s Disease 2024 ConferenceNeus Falgàs, Lucia Maure‐Blesa, Beau Ances, and 56 more authorsAlzheimer’s & Dementia, 2025The Down syndrome‐associated Alzheimer’s disease (DSAD) autosomal dominant Alzheimer’s disease (ADAD) 2024 Conference in Barcelona, convened under an Alzheimer’s Association International Society to Advance Alzheimer’s Research and Treatment (ISTAART) grant through the Down syndrome and Alzheimer’s disease (AD) Professional Interest Area (PIA), brought together global researchers to foster collaboration and knowledge exchange between the fields of DSAD and ADAD. This article provides a synthesis review of the conference proceedings, summarizing key discussions on biomarkers, natural history models, clinical trials, and ethical considerations in anti‐amyloid therapies. A total of 211 attendees from 16 countries joined the meeting. Global researchers presented on disease mechanisms, therapeutic developments, and patient care strategies. Discussions focused on challenges and opportunities unique to DSAD and ADAD. Experts emphasized the urgent need for tailored clinical trials for ADAD and DSAD and debated the safety and efficacy of anti‐amyloid treatments. Ethical considerations highlighted equitable access to therapies and the crucial role of patient and caregiver involvement. The conference highlighted the importance of inclusive research and collaboration across the genetic forms of AD. Biomarker research and natural history models developed in Down syndrome‐associated Alzheimer’s disease (DSAD) and autosomal dominant Alzheimer’s disease (ADAD) enable the prediction of disease progression not only for DSAD and ADAD, but also for sporadic Alzheimer’s disease (AD). ‐Collaboration and knowledge exchange among researchers across these genetic forms of AD will accelerate our understanding of the pathophysiology and advance preventive trials in DSAD and ADAD. ‐Tailored clinical trials for DSAD are urgently needed to address specific safety and efficacy concerns. ‐Inclusive research practices are crucial for advancing treatments and understanding of DSAD and ADAD.
-
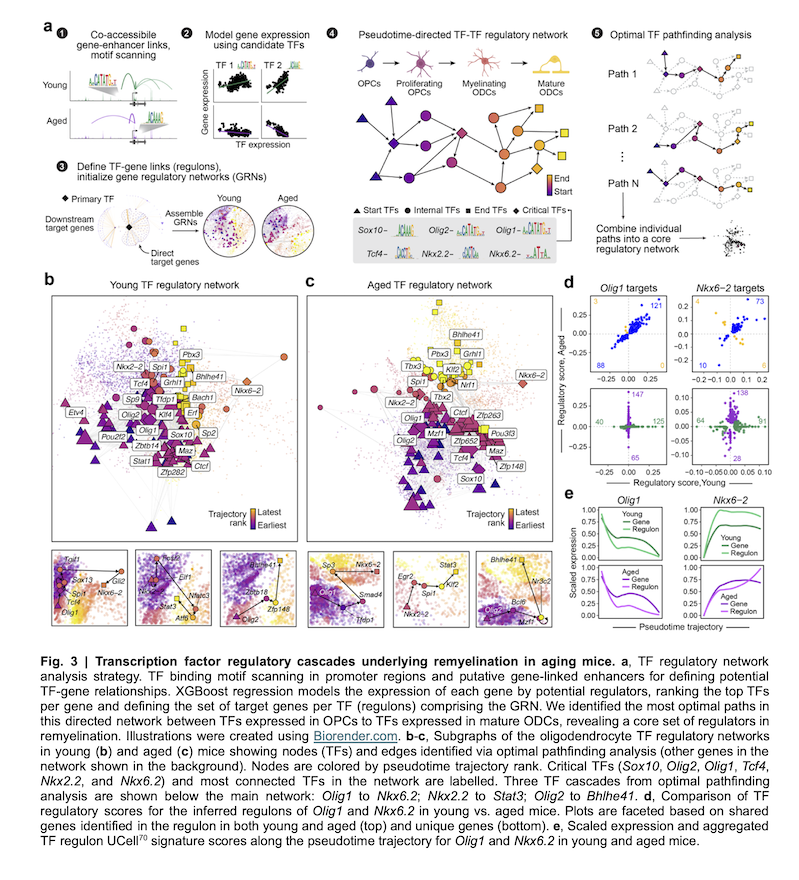 Dysregulation of transcription networks regulating oligodendrogenesis in age-related decline in CNS remyelinationPenelope Dimas, Samuel Morabito, Khalil S Rawji, and 12 more authors2025
Dysregulation of transcription networks regulating oligodendrogenesis in age-related decline in CNS remyelinationPenelope Dimas, Samuel Morabito, Khalil S Rawji, and 12 more authors2025In demyelinating diseases like multiple sclerosis (MS), efficient remyelination is critical for functional recovery. Remyelination efficiency declines with age, and is linked to progressive disability. The gene regulatory network underlying remyelination, and how it is altered with aging, remains unclear. Here we present a comparative single-nucleus RNA and ATAC sequencing analysis of remyelination in young and aged mice. We identified gene modules dynamically expressed throughout oligodendrocyte differentiation, revealing age-dependent changes in key processes related to myelination. Multi-omic analysis allowed us to map the regulatory network driving efficient remyelination within oligodendrocyte lineage cells in young mice. We highlight key transcription factors in the network dysregulated with age, and we describe similar dysregulations in MS lesions. Modifying the expression of these transcription factors in primary oligodendrocyte progenitor cells impacts differentiation. These findings provide a foundational understanding of this regenerative process in the context of aging and in chronic demyelinating diseases.
-
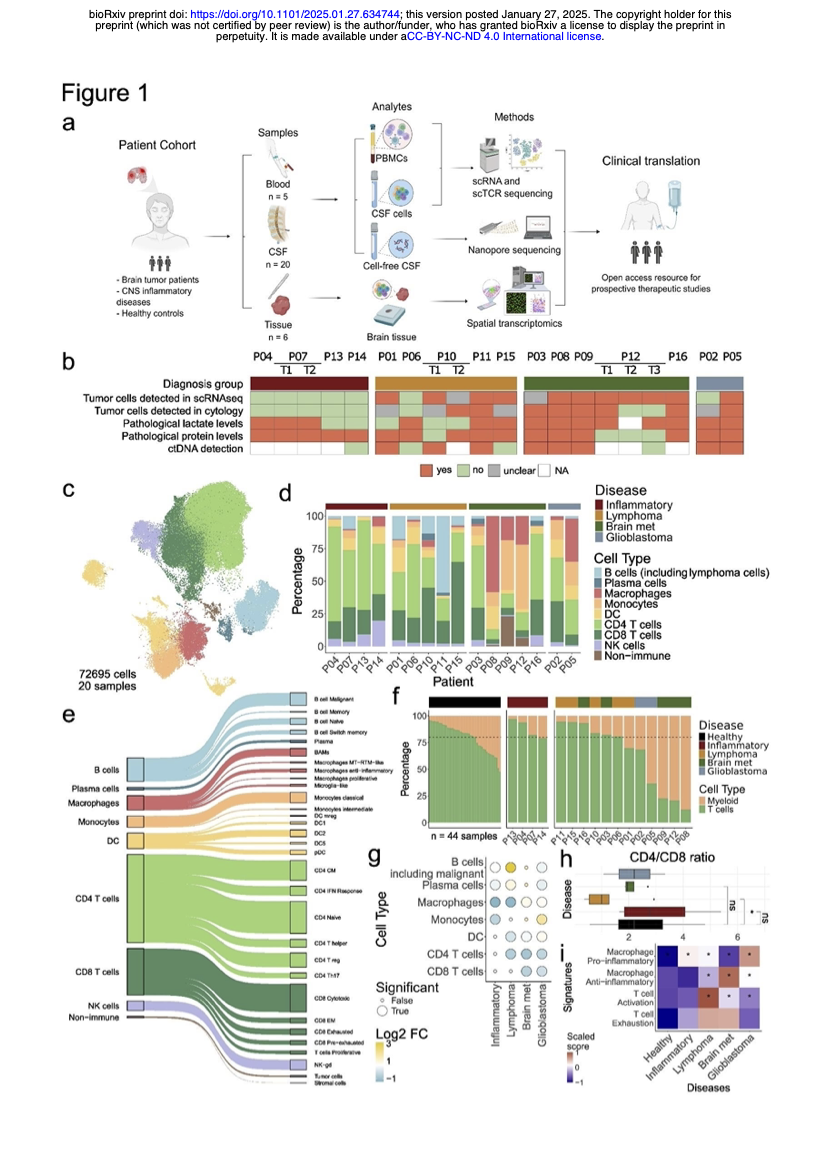 Decoding the immune response in leptomeningeal disease through single-cell sequencing of cerebrospinal fluidPaula Nieto, Svenja Klinsing, Ginevra Caratù, and 19 more authorsbioRxiv, 2025
Decoding the immune response in leptomeningeal disease through single-cell sequencing of cerebrospinal fluidPaula Nieto, Svenja Klinsing, Ginevra Caratù, and 19 more authorsbioRxiv, 2025Assessing anti-tumor immune responses and immune microenvironments in central nervous system (CNS) neoplasms, such as brain tumors and leptomeningeal disease (LMD), provides prognostic insights and predictive biomarkers. Liquid biopsy of the cerebrospinal fluid (CSF) represents a promising minimally-invasive approach, but its ability to reflect immune responses against tumors remains unclear. Here, we used single-cell sequencing of CSF cells and spatial transcriptomics of CNS lesions to compare and contrast LMD patients with CNS lymphoma (CNSL), glioblastoma (GB) and brain metastases (BrM), to neuroinflammatory CNS disorders. We identified disease-specific CSF environments, reflecting parenchymal tumor microenvironment features. CNSL showed robust T cell responses, while BrM and GB were dominated by both blood-derived and tissue-resident myeloid cells. Longitudinal CSF sampling unveiled mechanisms of disease progression and therapy resistance, highlighting the potential of CSF liquid biopsies for uncovering disease biology, discovering cellular biomarkers and developing personalized therapies for CNS neoplasms.
2024
-
 Spatial and single-nucleus transcriptomic analysis of genetic and sporadic forms of Alzheimer’s diseaseEmily Miyoshi*, Samuel Morabito*, Caden M Henningfield, and 24 more authorsNature Genetics, 2024
Spatial and single-nucleus transcriptomic analysis of genetic and sporadic forms of Alzheimer’s diseaseEmily Miyoshi*, Samuel Morabito*, Caden M Henningfield, and 24 more authorsNature Genetics, 2024The pathogenesis of Alzheimer’s disease (AD) depends on environmental and heritable factors, with its molecular etiology still unclear. Here we present a spatial transcriptomic (ST) and single-nucleus transcriptomic survey of late-onset sporadic AD and AD in Down syndrome (DSAD). Studying DSAD provides an opportunity to enhance our understanding of the AD transcriptome, potentially bridging the gap between genetic mouse models and sporadic AD. We identified transcriptomic changes that may underlie cortical layer-preferential pathology accumulation. Spatial co-expression network analyses revealed transient and regionally restricted disease processes, including a glial inflammatory program dysregulated in upper cortical layers and implicated in AD genetic risk and amyloid-associated processes. Cell–cell communication analysis further contextualized this gene program in dysregulated signaling networks. Finally, we generated ST data from an amyloid AD mouse model to identify cross-species amyloid-proximal transcriptomic changes with conformational context. Spatial and single-nucleus analyses in human postmortem Alzheimer’s disease (AD) brain tissues at early and late stages from individuals with and without Down syndrome, as well as in AD mouse models, show sex and species-specific phenotypic changes.
-
 Relapse to cocaine seeking is regulated by medial habenula NR4A2/NURR1 in miceJessica E. Childs*, Samuel Morabito*, Sudeshna Das, and 9 more authorsCell Reports, 2024
Relapse to cocaine seeking is regulated by medial habenula NR4A2/NURR1 in miceJessica E. Childs*, Samuel Morabito*, Sudeshna Das, and 9 more authorsCell Reports, 2024Drugs of abuse can persistently change the reward circuit in ways that contribute to relapse behavior, partly via mechanisms that regulate chromatin structure and function. Nuclear orphan receptor subfamily4 groupA member2 (NR4A2, also known as NURR1) is an important effector of histone deacetylase 3 (HDAC3)-dependent mechanisms in persistent memory processes and is highly expressed in the medial habenula (MHb), a region that regulates nicotine-associated behaviors. Here, expressing the Nr4a2 dominant negative (Nurr2c) in the MHb blocks reinstatement of cocaine seeking in mice. We use single-nucleus transcriptomics to characterize the molecular cascade following Nr4a2 manipulation, revealing changes in transcriptional networks related to addiction, neuroplasticity, and GABAergic and glutamatergic signaling. The network controlled by NR4A2 is characterized using a transcription factor regulatory network inference algorithm. These results identify the MHb as a pivotal regulator of relapse behavior and demonstrate the importance of NR4A2 as a key mechanism driving the MHb component of relapse.
-
 BIN1K358R suppresses glial response to plaques in mouse model of Alzheimer’s diseaseLaura Fernandez Garcia‐Agudo, Zechuan Shi, Ian F. Smith, and 24 more authorsAlzheimer’s & Dementia, 2024
BIN1K358R suppresses glial response to plaques in mouse model of Alzheimer’s diseaseLaura Fernandez Garcia‐Agudo, Zechuan Shi, Ian F. Smith, and 24 more authorsAlzheimer’s & Dementia, 2024The BIN1 coding variant rs138047593 (K358R) is linked to Late‐Onset Alzheimer’s Disease (LOAD) via targeted exome sequencing. To elucidate the functional consequences of this rare coding variant on brain amyloidosis and neuroinflammation, we generated BIN1K358R knock‐in mice using CRISPR/Cas9 technology. These mice were subsequently bred with 5xFAD transgenic mice, which serve as a model for Alzheimer’s pathology. The presence of the BIN1K358R variant leads to increased cerebral amyloid deposition, with a dampened response of astrocytes and oligodendrocytes, but not microglia, at both the cellular and transcriptional levels. This correlates with decreased neurofilament light chain in both plasma and brain tissue. Synaptic densities are significantly increased in both wild‐type and 5xFAD backgrounds homozygous for the BIN1K358R variant. The BIN1 K358R variant modulates amyloid pathology in 5xFAD mice, attenuates the astrocytic and oligodendrocytic responses to amyloid plaques, decreases damage markers, and elevates synaptic densities. BIN1 rs138047593 (K358R) coding variant is associated with increased risk of LOAD. BIN1 K358R variant increases amyloid plaque load in 12‐month‐old 5xFAD mice. BIN1 K358R variant dampens astrocytic and oligodendrocytic response to plaques. BIN1 K358R variant decreases neuronal damage in 5xFAD mice. BIN1 K358R upregulates synaptic densities and modulates synaptic transmission.
-
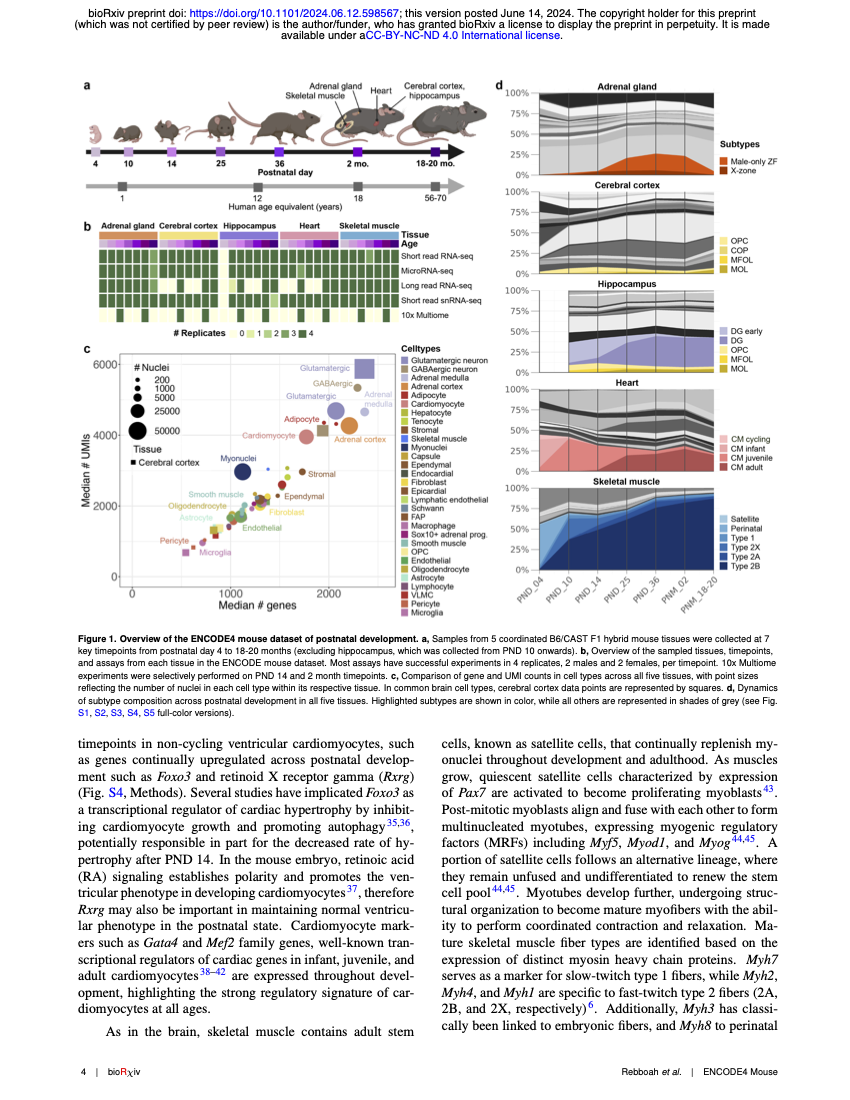 The ENCODE mouse postnatal developmental time course identifies regulatory programs of cell types and cell statesElisabeth Rebboah, Narges Rezaie, Brian A Williams, and 14 more authors2024
The ENCODE mouse postnatal developmental time course identifies regulatory programs of cell types and cell statesElisabeth Rebboah, Narges Rezaie, Brian A Williams, and 14 more authors2024Postnatal genomic regulation significantly influences tissue and organ maturation but is under-studied relative to existing genomic catalogs of adult tissues or prenatal development in mouse. The ENCODE4 consortium generated the first comprehensive single-nucleus resource of postnatal regulatory events across a diverse set of mouse tissues. The collection spans seven postnatal time points, mirroring human development from childhood to adulthood, and encompasses five core tissues. We identified 30 cell types, further subdivided into 69 subtypes and cell states across adrenal gland, left cerebral cortex, hippocampus, heart, and gastrocnemius muscle. Our annotations cover both known and novel cell differentiation dynamics ranging from early hippocampal neurogenesis to a new sex-specific adrenal gland population during puberty. We used an ensemble Latent Dirichlet Allocation strategy with a curated vocabulary of 2,701 regulatory genes to identify regulatory "topics," each of which is a gene vector, linked to cell type differentiation, subtype specialization, and transitions between cell states. We find recurrent regulatory topics in tissue-resident macrophages, neural cell types, endothelial cells across multiple tissues, and cycling cells of the adrenal gland and heart. Cell-type-specific topics are enriched in transcription factors and microRNA host genes, while chromatin regulators dominate mitosis topics. Corresponding chromatin accessibility data reveal dynamic and sex-specific regulatory elements, with enriched motifs matching transcription factors in regulatory topics. Together, these analyses identify both tissue-specific and common regulatory programs in postnatal development across multiple tissues through the lens of the factors regulating transcription.
2023
-
 hdWGCNA identifies co-expression networks in high-dimensional transcriptomics dataSamuel Morabito, Fairlie Reese, Negin Rahimzadeh, and 2 more authorsCell Reports Methods, 2023
hdWGCNA identifies co-expression networks in high-dimensional transcriptomics dataSamuel Morabito, Fairlie Reese, Negin Rahimzadeh, and 2 more authorsCell Reports Methods, 2023Biological systems are immensely complex, organized into a multi-scale hierarchy of functional units based on tightly regulated interactions between distinct molecules, cells, organs, and organisms. While experimental methods enable transcriptome-wide measurements across millions of cells, popular bioinformatic tools do not support systems-level analysis. Here we present hdWGCNA, a comprehensive framework for analyzing co-expression networks in high-dimensional transcriptomics data such as single-cell and spatial RNA sequencing (RNA-seq). hdWGCNA provides functions for network inference, gene module identification, gene enrichment analysis, statistical tests, and data visualization. Beyond conventional single-cell RNA-seq, hdWGCNA is capable of performing isoform-level network analysis using long-read single-cell data. We showcase hdWGCNA using data from autism spectrum disorder and Alzheimer’s disease brain samples, identifying disease-relevant co-expression network modules. hdWGCNA is directly compatible with Seurat, a widely used R package for single-cell and spatial transcriptomics analysis, and we demonstrate the scalability of hdWGCNA by analyzing a dataset containing nearly 1 million cells
-
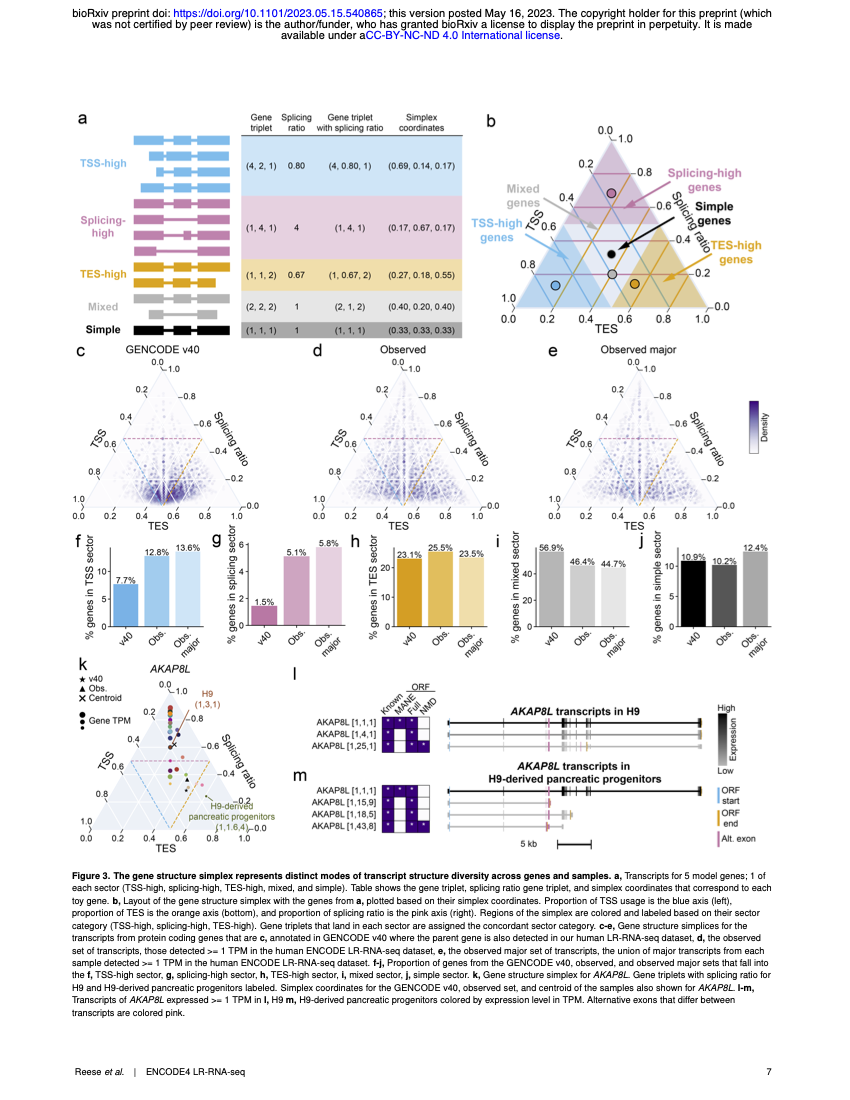 The ENCODE4 long-read RNA-seq collection reveals distinct classes of transcript structure diversityFairlie Reese, Brian Williams, Gabriela Balderrama-Gutierrez, and 42 more authorsbioRxiv, 2023
The ENCODE4 long-read RNA-seq collection reveals distinct classes of transcript structure diversityFairlie Reese, Brian Williams, Gabriela Balderrama-Gutierrez, and 42 more authorsbioRxiv, 2023The majority of mammalian genes encode multiple transcript isoforms that result from differential promoter use, changes in exonic splicing, and alternative 3′ end choice. Detecting and quantifying transcript isoforms across tissues, cell types, and species has been extremely challenging because transcripts are much longer than the short reads normally used for RNA-seq. By contrast, long-read RNA-seq (LR-RNA-seq) gives the complete structure of most transcripts. We sequenced 264 LR-RNA-seq PacBio libraries totaling over 1 billion circular consensus reads (CCS) for 81 unique human and mouse samples. We detect at least one full-length transcript from 87.7% of annotated human protein coding genes and a total of \textbackslashtextasciitilde200,000 full-length transcripts, \textbackslashtextasciitilde40% of which have novel exon junction chains. To capture and compute on the three sources of transcript structure diversity, we introduce a gene and transcript annotation framework that uses triplets representing the transcript start site, exon junction chain, and transcript end site of each transcript. Using triplets in a simplex representation demonstrates how promoter selection, splice pattern, and 3′ processing are deployed across human tissues, with nearly half of multitranscript protein coding genes showing a clear bias toward one of the three diversity mechanisms. Evaluated across samples, the predominantly expressed transcript changes for 74% of protein coding genes. In evolution, the human and mouse transcriptomes are globally similar in types of transcript structure diversity, yet among individual orthologous gene pairs, more than half (57.8%) show substantial differences in mechanism of diversification in matching tissues. This initial large-scale survey of human and mouse long-read transcriptomes provides a foundation for further analyses of alternative transcript usage, and is complemented by short-read and microRNA data on the same samples and by epigenome data elsewhere in the ENCODE4 collection.
-
 The Type 2 Diabetes Knowledge Portal: An open access genetic resource dedicated to type 2 diabetes and related traitsMaria C. Costanzo, Marcin von Grotthuss, Jeffrey Massung, and 189 more authorsCell Metabolism, 2023
The Type 2 Diabetes Knowledge Portal: An open access genetic resource dedicated to type 2 diabetes and related traitsMaria C. Costanzo, Marcin von Grotthuss, Jeffrey Massung, and 189 more authorsCell Metabolism, 2023Associations between human genetic variation and clinical phenotypes have become a foundation of biomedical research. Most repositories of these data seek to be disease-agnostic and therefore lack disease-focused views. The Type 2 Diabetes Knowledge Portal (T2DKP) is a public resource of genetic datasets and genomic annotations dedicated to type 2 diabetes (T2D) and related traits. Here, we seek to make the T2DKP more accessible to prospective users and more useful to existing users. First, we evaluate the T2DKP’s comprehensiveness by comparing its datasets with those of other repositories. Second, we describe how researchers unfamiliar with human genetic data can begin using and correctly interpreting them via the T2DKP. Third, we describe how existing users can extend their current workflows to use the full suite of tools offered by the T2DKP. We finally discuss the lessons offered by the T2DKP toward the goal of democratizing access to complex disease genetic results.
2022
-
 Molecular signatures underlying neurofibrillary tangle susceptibility in Alzheimer’s diseaseMarcos Otero-Garcia, Sameehan U. Mahajani, Debia Wakhloo, and 14 more authorsNeuron, 2022
Molecular signatures underlying neurofibrillary tangle susceptibility in Alzheimer’s diseaseMarcos Otero-Garcia, Sameehan U. Mahajani, Debia Wakhloo, and 14 more authorsNeuron, 2022Tau aggregation in neurofibrillary tangles (NFTs) is closely associated with neurodegeneration and cognitive decline in Alzheimer’s disease (AD). However, the molecular signatures that distinguish between aggregation-prone and aggregation-resistant cell states are unknown. We developed methods for the high-throughput isolation and transcriptome profiling of single somas with NFTs from the human AD brain, quantified the susceptibility of 20 neocortical subtypes for NFT formation and death, and identified both shared and cell-type-specific signatures. NFT-bearing neurons shared a marked upregulation of synaptic transmission-related genes, including a core set of 63 genes enriched for synaptic vesicle cycling. Oxidative phosphorylation and mitochondrial dysfunction were highly cell-type dependent. Apoptosis was only modestly enriched, and the susceptibilities of NFT-bearing and NFT-free neurons for death were highly similar. Our analysis suggests that NFTs represent cell-type-specific responses to stress and synaptic dysfunction. We provide a resource for biomarker discovery and the investigation of tau-dependent and tau-independent mechanisms of neurodegeneration.
-
 Absence of microglia promotes diverse pathologies and early lethality in Alzheimer’s disease miceSepideh Kiani Shabestari, Samuel Morabito, Emma Pascal Danhash, and 18 more authorsCell Reports, 2022
Absence of microglia promotes diverse pathologies and early lethality in Alzheimer’s disease miceSepideh Kiani Shabestari, Samuel Morabito, Emma Pascal Danhash, and 18 more authorsCell Reports, 2022Microglia are strongly implicated in the development and progression of Alzheimer’s disease (AD), yet their impact on pathology and lifespan remains unclear. Here we utilize a CSF1R hypomorphic mouse to generate a model of AD that genetically lacks microglia. The resulting microglial-deficient mice exhibit a profound shift from parenchymal amyloid plaques to cerebral amyloid angiopathy (CAA), which is accompanied by numerous transcriptional changes, greatly increased brain calcification and hemorrhages, and premature lethality. Remarkably, a single injection of wild-type microglia into adult mice repopulates the microglial niche and prevents each of these pathological changes. Taken together, these results indicate the protective functions of microglia in reducing CAA, blood-brain barrier dysfunction, and brain calcification. To further understand the clinical implications of these findings, human AD tissue and iPSC-microglia were examined, providing evidence that microglia phagocytose calcium crystals, and this process is impaired by loss of the AD risk gene, TREM2.
-
 Protocol for single-nucleus ATAC sequencing and bioinformatic analysis in frozen human brain tissueZechuan Shi*, Sudeshna Das*, Samuel Morabito, and 2 more authorsSTAR Protocols, 2022
Protocol for single-nucleus ATAC sequencing and bioinformatic analysis in frozen human brain tissueZechuan Shi*, Sudeshna Das*, Samuel Morabito, and 2 more authorsSTAR Protocols, 2022Single-nucleus ATAC sequencing (snATAC-seq) employs a hyperactive Tn5 transposase to gain precise information about the cis-regulatory elements in specific cell types. However, the standard protocol of snATAC-seq is not optimized for all tissues, including the brain. Here, we present a modified protocol for single-nuclei isolation from postmortem frozen human brain tissue, followed by snATAC-seq library preparation and sequencing. We also describe an integrated bioinformatics analysis pipeline using an R package (ArchRtoSignac) to robustly analyze snATAC-seq data. For complete details on the use and execution of this protocol, please refer to Morabito et al. (2021).
-
 Cortical diurnal rhythms remain intact with microglial depletionRocio A. Barahona, Samuel Morabito, Vivek Swarup, and 1 more authorScientific Reports, 2022
Cortical diurnal rhythms remain intact with microglial depletionRocio A. Barahona, Samuel Morabito, Vivek Swarup, and 1 more authorScientific Reports, 2022Microglia are subject to change in tandem with the endogenously generated biological oscillations known as our circadian rhythm. Studies have shown microglia harbor an intrinsic molecular clock which regulates diurnal changes in morphology and influences inflammatory responses. In the adult brain, microglia play an important role in the regulation of condensed extracellular matrix structures called perineuronal nets (PNNs), and it has been suggested that PNNs are also regulated in a circadian and diurnal manner. We sought to determine whether microglia mediate the diurnal regulation of PNNs via CSF1R inhibitor dependent microglial depletion in C57BL/6J mice, and how the absence of microglia might affect cortical diurnal gene expression rhythms. While we observe diurnal differences in microglial morphology, where microglia are most ramified at the onset of the dark phase, we do not find diurnal differences in PNN intensity. However, PNN intensity increases across many brain regions in the absence of microglia, supporting a role for microglia in the regulation of PNNs. Here, we also show that cortical diurnal gene expression rhythms are intact, with no cycling gene changes without microglia. These findings demonstrate a role for microglia in the maintenance of PNNs, but not in the maintenance of diurnal rhythms.
2021
-
 Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s diseaseSamuel Morabito*, Emily Miyoshi*, Neethu Michael, and 7 more authorsNature Genetics, 2021
Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s diseaseSamuel Morabito*, Emily Miyoshi*, Neethu Michael, and 7 more authorsNature Genetics, 2021The gene-regulatory landscape of the brain is highly dynamic in health and disease, coordinating a menagerie of biological processes across distinct cell types. Here, we present a multi-omic single-nucleus study of 191,890 nuclei in late-stage Alzheimer’s disease (AD), accessible through our web portal, profiling chromatin accessibility and gene expression in the same biological samples and uncovering vast cellular heterogeneity. We identified cell-type-specific, disease-associated candidate cis-regulatory elements and their candidate target genes, including an oligodendrocyte-associated regulatory module containing links to APOE and CLU. We describe cis-regulatory relationships in specific cell types at a subset of AD risk loci defined by genome-wide association studies, demonstrating the utility of this multi-omic single-nucleus approach. Trajectory analysis of glial populations identified disease-relevant transcription factors, such as SREBF1, and their regulatory targets. Finally, we introduce single-nucleus consensus weighted gene coexpression analysis, a coexpression network analysis strategy robust to sparse single-cell data, and perform a systems-level analysis of the AD transcriptome.
-
 Systems biology approaches to unravel the molecular and genetic architecture of Alzheimer’s disease and related tauopathiesEmily Miyoshi*, Samuel Morabito*, and Vivek SwarupNeurobiology of Disease, 2021
Systems biology approaches to unravel the molecular and genetic architecture of Alzheimer’s disease and related tauopathiesEmily Miyoshi*, Samuel Morabito*, and Vivek SwarupNeurobiology of Disease, 2021Over the years, genetic studies have identified multiple genetic risk variants associated with neurodegenerative disorders and helped reveal new biological pathways and genes of interest. However, genetic risk variants commonly reside in non-coding regions and may regulate distant genes rather than the nearest gene, as well as a gene’s interaction partners in biological networks. Systems biology and functional genomics approaches provide the framework to unravel the functional significance of genetic risk variants in disease. In this review, we summarize the genetic and transcriptomic studies of Alzheimer’s disease and related tauopathies and focus on the advantages of performing systems-level analyses to interrogate the biological pathways underlying neurodegeneration. Finally, we highlight new avenues of multi-omics analysis with single-cell approaches, which provides unparalleled opportunities to systematically explore cellular heterogeneity, and present an example of how to integrate publicly available single-cell datasets. Systems-level analysis has illuminated the function of many disease risk genes, but much work remains to study tauopathies and to understand spatiotemporal gene expression changes of specific cell types.
2020
-
 Integrative genomics approach identifies conserved transcriptomic networks in Alzheimer’s diseaseSamuel Morabito, Emily Miyoshi, Neethu Michael, and 1 more authorHuman Molecular Genetics, Aug 2020
Integrative genomics approach identifies conserved transcriptomic networks in Alzheimer’s diseaseSamuel Morabito, Emily Miyoshi, Neethu Michael, and 1 more authorHuman Molecular Genetics, Aug 2020Alzheimer’s disease (AD) is a devastating neurological disorder characterized by changes in cell-type proportions and consequently marked alterations of the transcriptome. Here we use a data-driven systems biology meta-analytical approach across three human ad cohorts, encompassing six cortical brain regions, and integrate with multi-scale datasets comprising of DNA methylation, histone acetylation, transcriptome- and genome-wide association studies, and quantitative trait loci to further characterize the genetic architecture of ad. We perform co-expression network analysis across more than twelve hundred human brain samples, identifying robust ad-associated dysregulation of the transcriptome, unaltered in normal human aging. We assess the cell-type specificity of ad gene co-expression changes and estimate cell-type proportion changes in human ad by integrating co-expression modules with single-cell transcriptome data generated from 27 321 nuclei from human postmortem prefrontal cortical tissue. We also show that genetic variants of ad are enriched in a microglial ad-associated module and identify key transcription factors regulating co-expressed modules. Additionally, we validate our results in multiple published human ad gene expression datasets, which can be easily accessed using our online resource (https://swaruplab.bio.uci.edu/consensusAD).
2019
-
 Single-cell landscape in mammary epithelium reveals bipotent-like cells associated with breast cancer risk and outcomeWeiyan Chen, Samuel J. Morabito, Kai Kessenbrock, and 3 more authorsCommunications Biology, Dec 2019
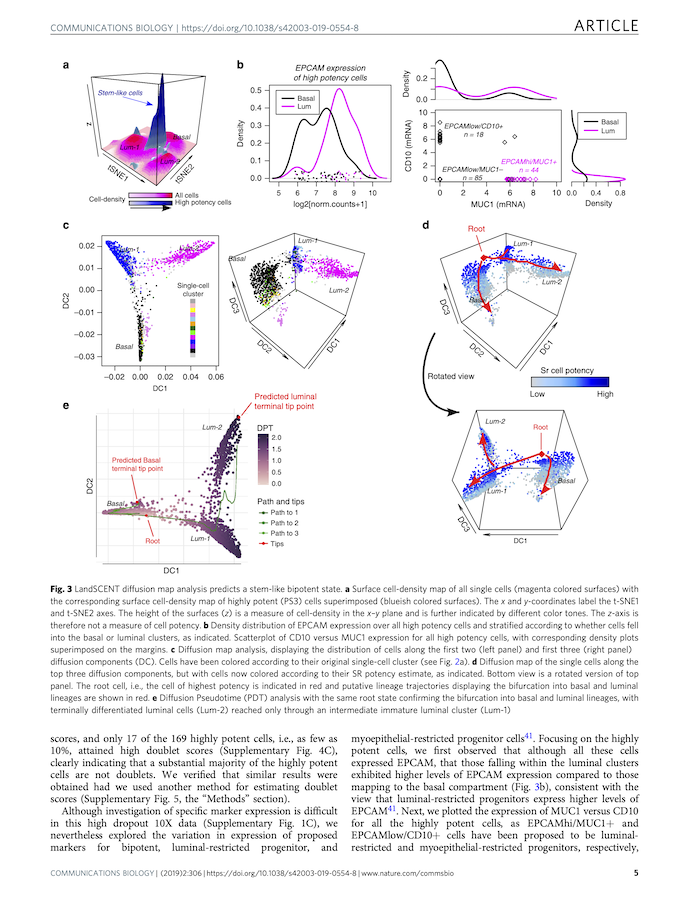
Single-cell landscape in mammary epithelium reveals bipotent-like cells associated with breast cancer risk and outcomeWeiyan Chen, Samuel J. Morabito, Kai Kessenbrock, and 3 more authorsCommunications Biology, Dec 2019Adult stem-cells may serve as the cell-of-origin for cancer, yet their unbiased identification in single cell RNA sequencing data is challenging due to the high dropout rate. In the case of breast, the existence of a bipotent stem-like state is also controversial. Here we apply a marker-free algorithm to scRNA-Seq data from the human mammary epithelium, revealing a high-potency cell-state enriched for an independent mammary stem-cell expression module. We validate this stem-like state in independent scRNA-Seq data. Our algorithm further predicts that the stem-like state is bipotent, a prediction we are able to validate using FACS sorted bulk expression data. The bipotent stem-like state correlates with clinical outcome in basal breast cancer and is characterized by overexpression of YBX1 and ENO1, two modulators of basal breast cancer risk. This study illustrates the power of a marker-free computational framework to identify a novel bipotent stem-like state in the mammary epithelium.